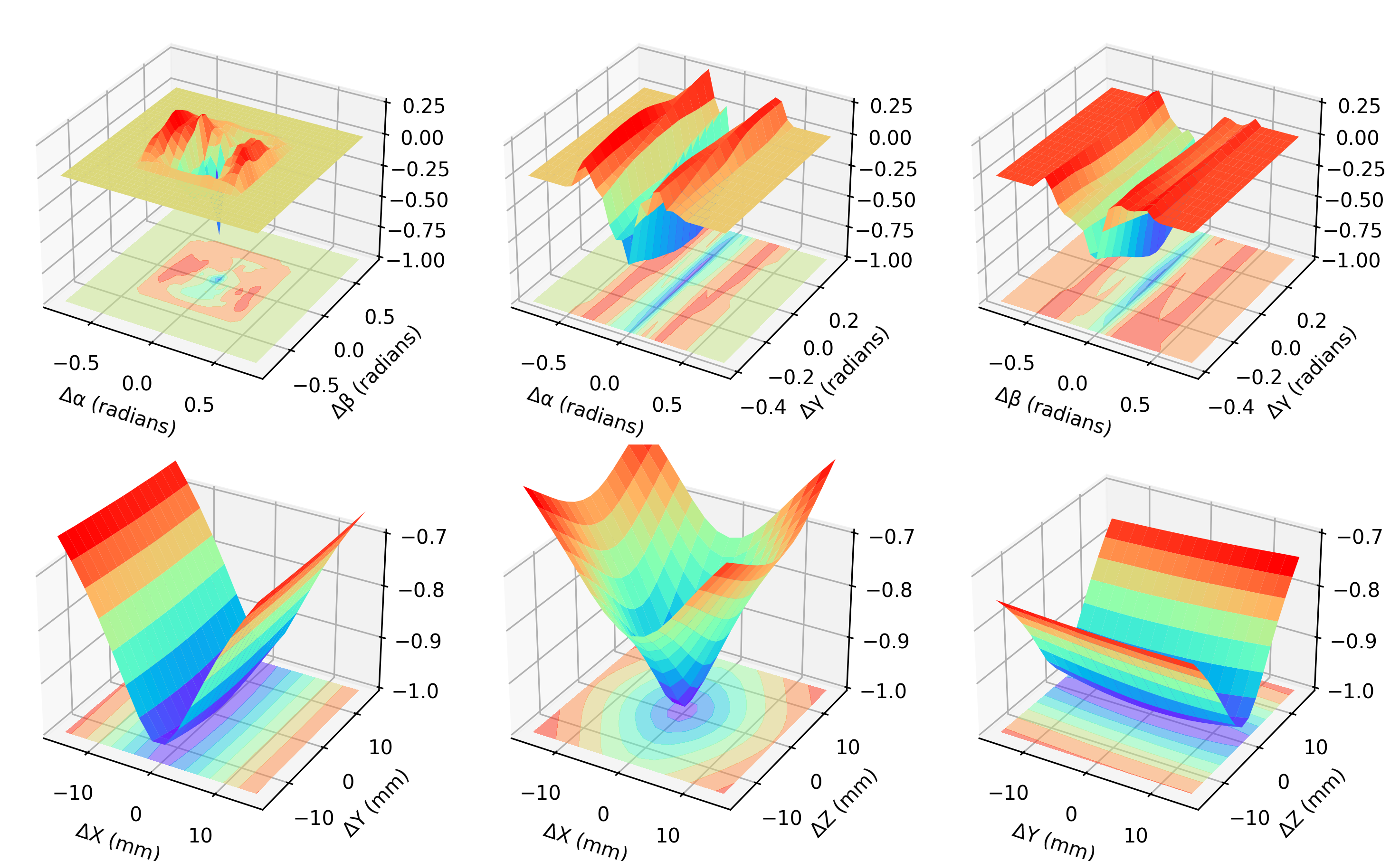
### NCC for the XYZs
xs = torch.arange(-15.0, 16.0, step=2)
ys = torch.arange(-15.0, 16.0, step=2)
zs = torch.arange(-15.0, 16.0, step=2)
# Get coordinate-wise correlations
xy_corrs = []
for x in tqdm(xs, desc="XY", ncols=50):
for y in ys:
xcorr = get_metric(alpha, beta, gamma, bx + x, by + y, bz)
xy_corrs.append(-xcorr)
XY = torch.tensor(xy_corrs).reshape(len(xs), len(ys))
xz_corrs = []
for x in tqdm(xs, desc="XZ", ncols=50):
for z in zs:
xcorr = get_metric(alpha, beta, gamma, bx + x, by, bz + z)
xz_corrs.append(-xcorr)
XZ = torch.tensor(xz_corrs).reshape(len(xs), len(zs))
yz_corrs = []
for y in tqdm(ys, desc="YZ", ncols=50):
for z in zs:
xcorr = get_metric(alpha, beta, gamma, bx, by + y, bz + z)
yz_corrs.append(-xcorr)
YZ = torch.tensor(yz_corrs).reshape(len(ys), len(zs))
### NCC for the angles
a_angles = torch.arange(-torch.pi / 4, torch.pi / 4, step=0.05)
b_angles = torch.arange(-torch.pi / 4, torch.pi / 4, step=0.05)
g_angles = torch.arange(-torch.pi / 8, torch.pi / 8, step=0.05)
# Get coordinate-wise correlations
tp_corrs = []
for t in tqdm(a_angles, desc="αβ", ncols=50):
for p in b_angles:
xcorr = get_metric(alpha + t, beta + p, gamma, bx, by, bz)
tp_corrs.append(-xcorr)
TP = torch.tensor(tp_corrs).reshape(len(a_angles), len(b_angles))
tg_corrs = []
for t in tqdm(a_angles, desc="αγ", ncols=50):
for g in g_angles:
xcorr = get_metric(alpha + t, beta, gamma + g, bx, by, bz)
tg_corrs.append(-xcorr)
TG = torch.tensor(tg_corrs).reshape(len(a_angles), len(g_angles))
pg_corrs = []
for p in tqdm(b_angles, desc="βγ", ncols=50):
for g in g_angles:
xcorr = get_metric(alpha, beta + p, gamma + g, bx, by, bz)
pg_corrs.append(-xcorr)
PG = torch.tensor(pg_corrs).reshape(len(b_angles), len(g_angles))
### Make the plots
# XYZ
xyx, xyy = torch.meshgrid(xs, ys, indexing="ij")
xzx, xzz = torch.meshgrid(xs, zs, indexing="ij")
yzy, yzz = torch.meshgrid(ys, zs, indexing="ij")
fig = plt.figure(figsize=3 * plt.figaspect(1.2 / 1), dpi=300)
ax = fig.add_subplot(1, 3, 1, projection="3d")
ax.contourf(xyx, xyy, XY, zdir="z", offset=-1, cmap=plt.get_cmap("rainbow"), alpha=0.5)
ax.plot_surface(xyx, xyy, XY, rstride=1, cstride=1, cmap=plt.get_cmap("rainbow"))
ax.set_xlabel("ΔX (mm)")
ax.set_ylabel("ΔY (mm)")
ax.set_zlim3d(-1.0, -0.6)
ax = fig.add_subplot(1, 3, 2, projection="3d")
ax.contourf(xzx, xzz, XZ, zdir="z", offset=-1, cmap=plt.get_cmap("rainbow"), alpha=0.5)
ax.plot_surface(xzx, xzz, XZ, rstride=1, cstride=1, cmap=plt.get_cmap("rainbow"))
ax.set_xlabel("ΔX (mm)")
ax.set_ylabel("ΔZ (mm)")
ax.set_zlim3d(-1.0, -0.6)
ax = fig.add_subplot(1, 3, 3, projection="3d")
ax.contourf(yzy, yzz, YZ, zdir="z", offset=-1, cmap=plt.get_cmap("rainbow"), alpha=0.5)
ax.plot_surface(yzy, yzz, YZ, rstride=1, cstride=1, cmap=plt.get_cmap("rainbow"))
ax.set_xlabel("ΔY (mm)")
ax.set_ylabel("ΔZ (mm)")
ax.set_zlim3d(-1.0, -0.6)
# Angles
xyx, xyy = torch.meshgrid(a_angles, b_angles, indexing="ij")
xzx, xzz = torch.meshgrid(a_angles, g_angles, indexing="ij")
yzy, yzz = torch.meshgrid(b_angles, g_angles, indexing="ij")
ax = fig.add_subplot(2, 3, 1, projection="3d")
ax.contourf(xyx, xyy, TP, zdir="z", offset=-1, cmap=plt.get_cmap("rainbow"), alpha=0.5)
ax.plot_surface(xyx, xyy, TP, rstride=1, cstride=1, cmap=plt.get_cmap("rainbow"))
ax.set_xlabel("Δα (radians)")
ax.set_ylabel("Δβ (radians)")
ax.set_zlim3d(-1.0, -0.25)
ax = fig.add_subplot(2, 3, 2, projection="3d")
ax.contourf(xzx, xzz, TG, zdir="z", offset=-1, cmap=plt.get_cmap("rainbow"), alpha=0.5)
ax.plot_surface(xzx, xzz, TG, rstride=1, cstride=1, cmap=plt.get_cmap("rainbow"))
ax.set_xlabel("Δα (radians)")
ax.set_ylabel("Δγ (radians)")
ax.set_zlim3d(-1.0, -0.25)
ax = fig.add_subplot(2, 3, 3, projection="3d")
ax.contourf(yzy, yzz, PG, zdir="z", offset=-1, cmap=plt.get_cmap("rainbow"), alpha=0.5)
ax.plot_surface(yzy, yzz, PG, rstride=1, cstride=1, cmap=plt.get_cmap("rainbow"))
ax.set_xlabel("Δβ (radians)")
ax.set_ylabel("Δγ (radians)")
ax.set_zlim3d(-1.0, -0.25)
plt.show()