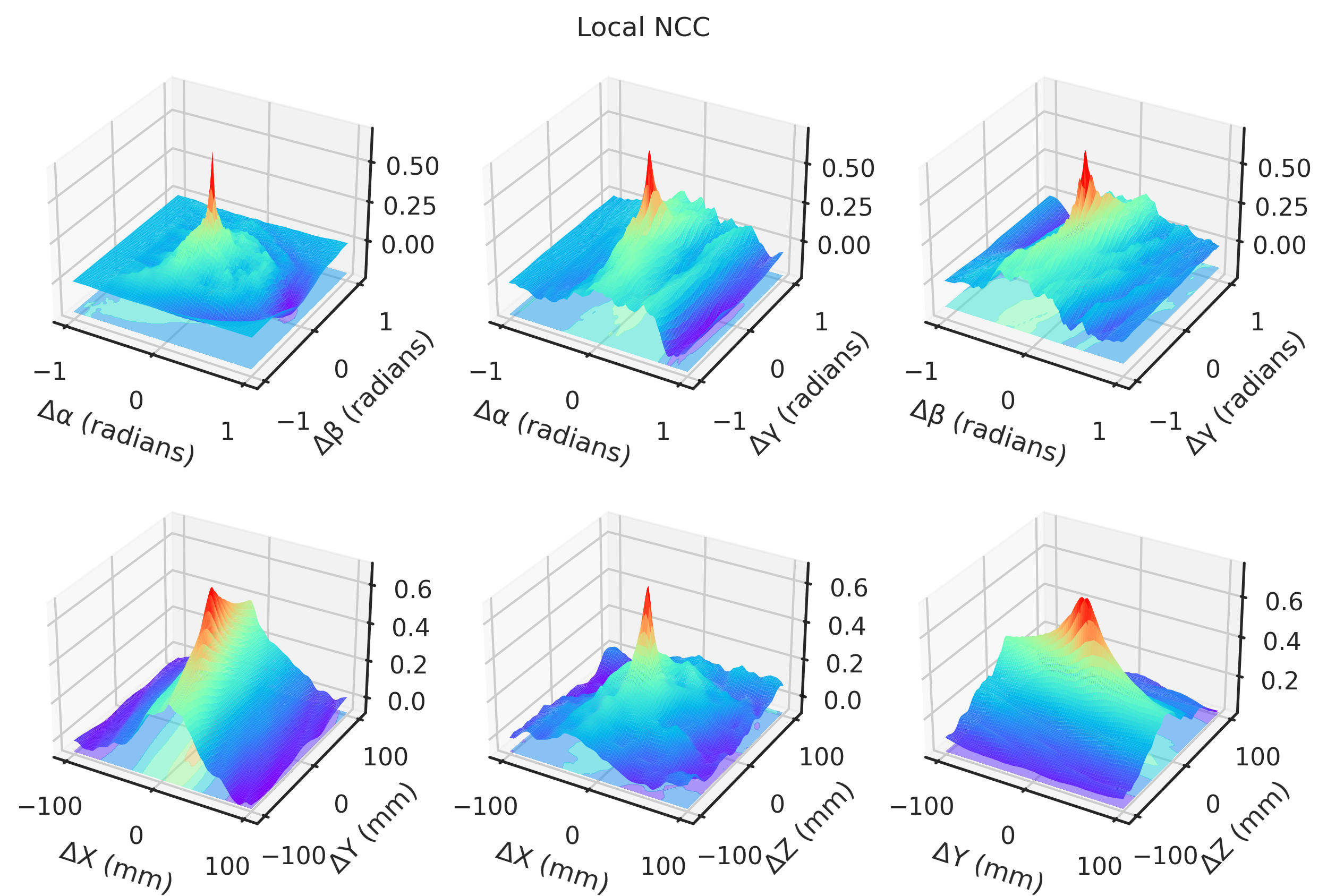
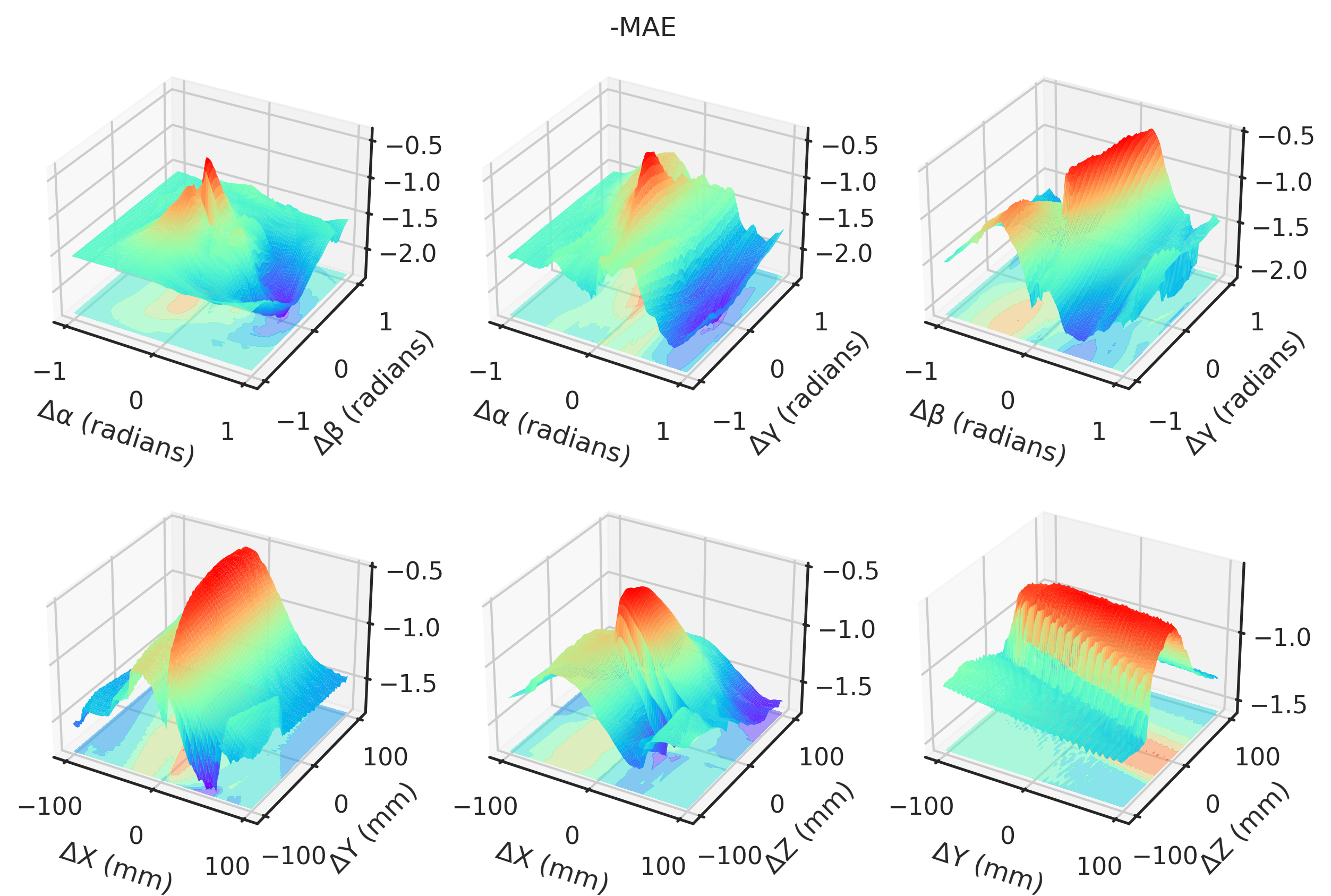
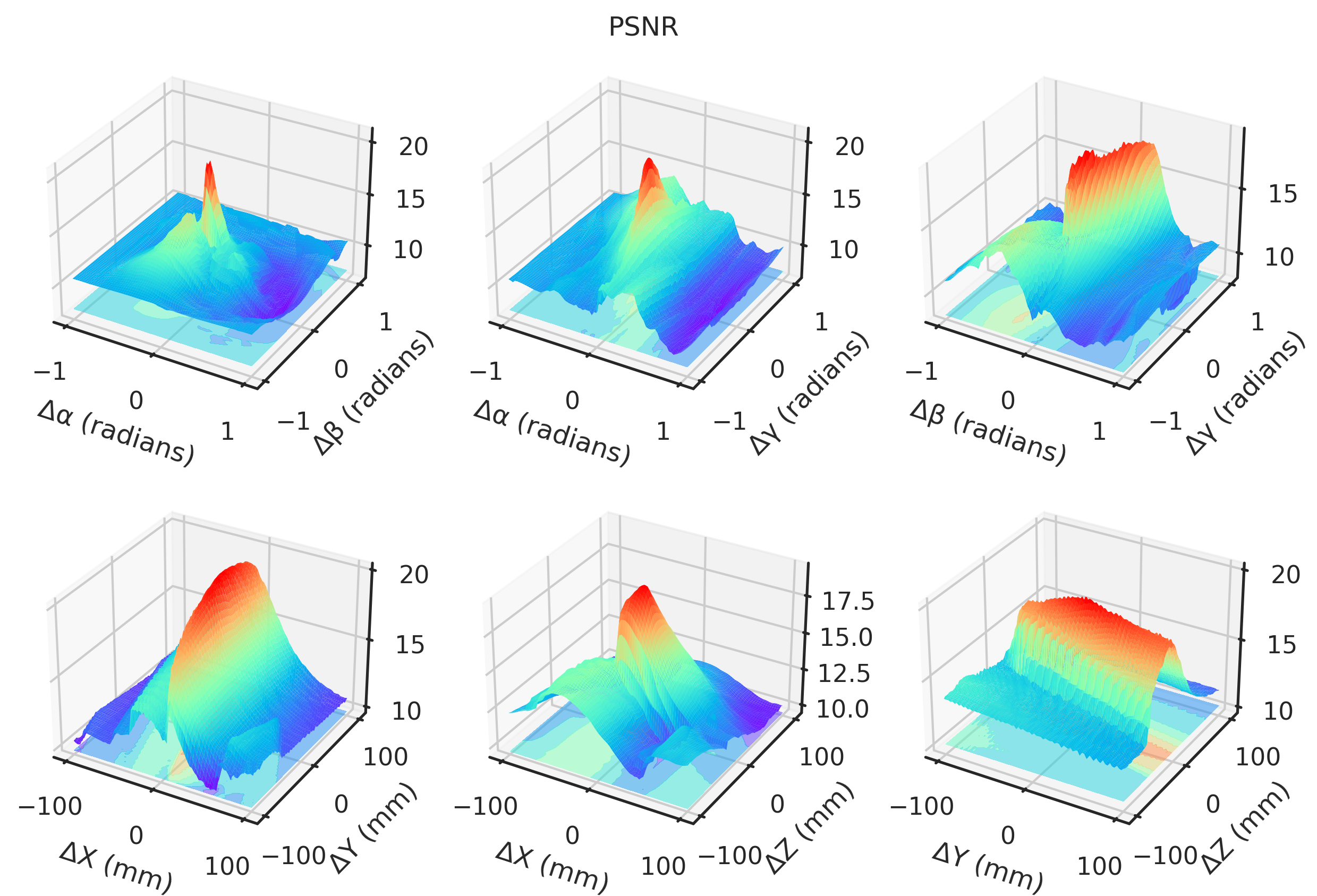
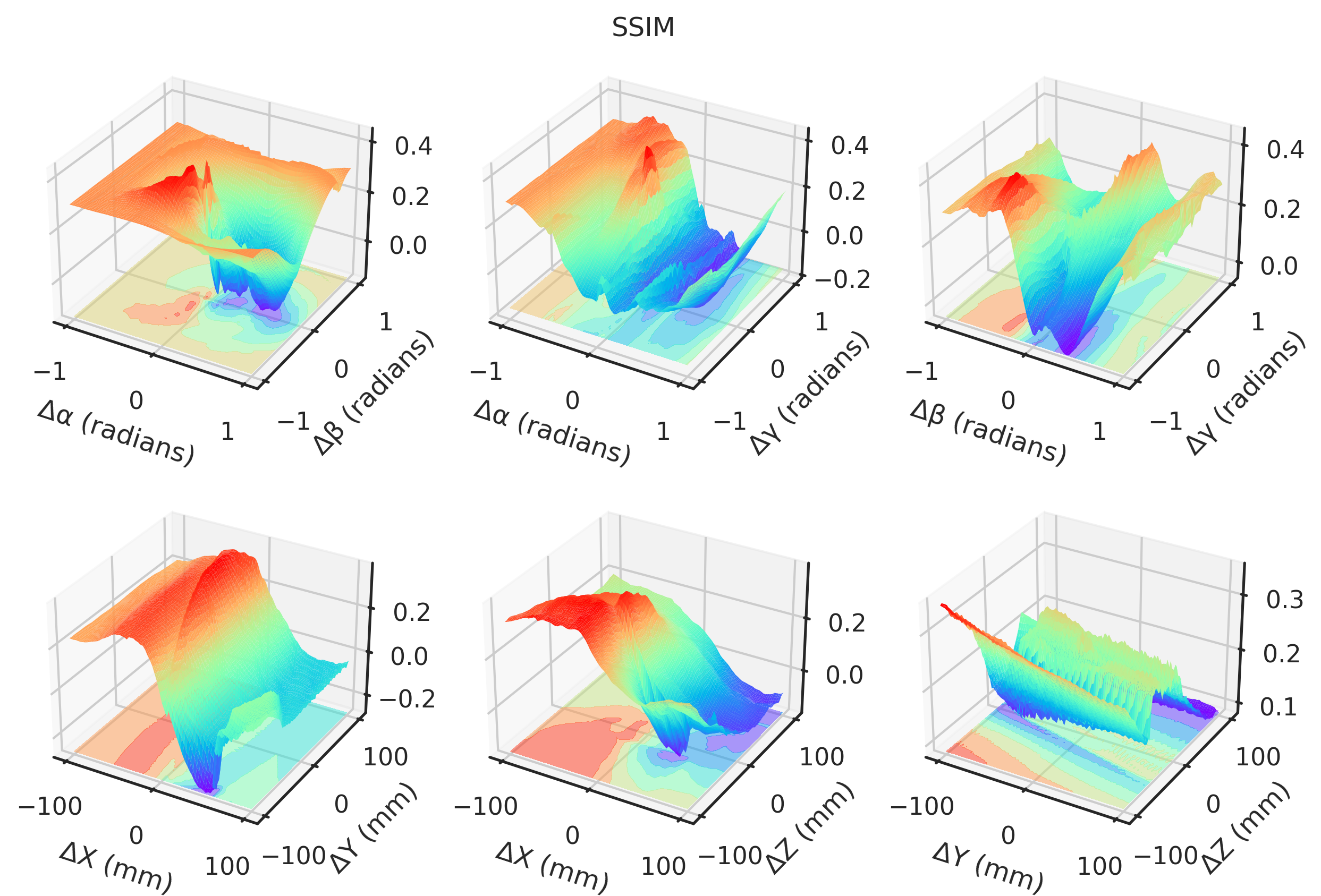
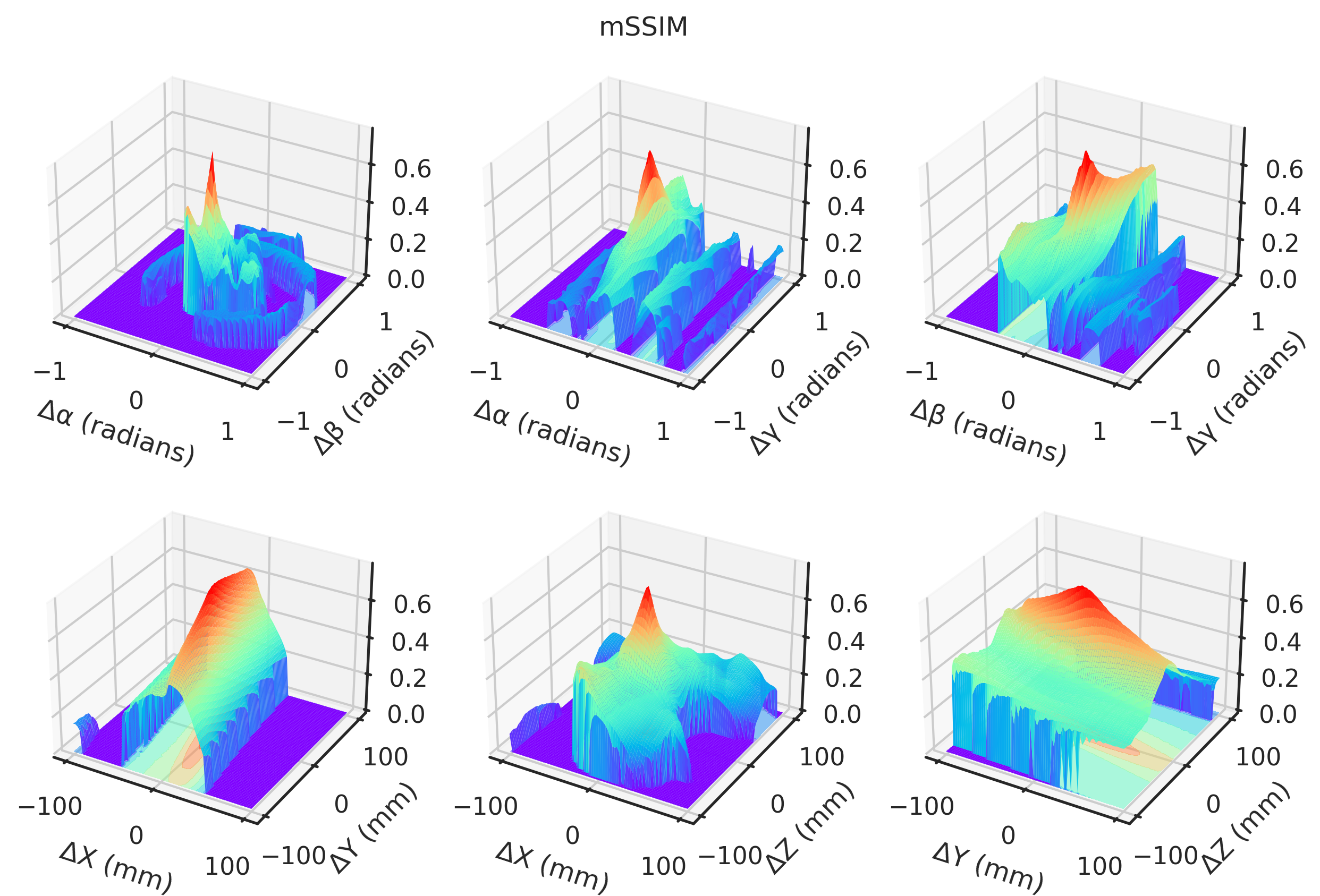
= "notebook" , style= "ticks" )def plot(idx, zmin= None , zmax= None ):if idx == 2 or idx == 3 := - 1 else := 1 ### 3D = plt.figure(figsize= (10 , 6.5 ), dpi= 300 )= []# Angles = torch.meshgrid(t_angles, p_angles, indexing= "ij" )= torch.meshgrid(t_angles, g_angles, indexing= "ij" )= torch.meshgrid(p_angles, g_angles, indexing= "ij" )= fig.add_subplot(2 , 3 , 1 , projection= "3d" )* TP[..., idx].numpy(),= "z" ,= (multiplier * TP[..., idx]).min (),= plt.get_cmap("rainbow" ),= 0.5 ,* TP[..., idx].numpy(),= 1 ,= 1 ,= plt.get_cmap("rainbow" ),= 0.0 ,"Δα (radians)" )"Δβ (radians)" )= fig.add_subplot(2 , 3 , 2 , projection= "3d" )"Gradient NCC" ,"Local NCC" ,"-MAE" ,"-MSE" ,"Global NCC" ,"PSNR" ,"SSIM" ,"mNCC" ,"mSSIM" ,* TG[..., idx].numpy(),= "z" ,= (multiplier * TG[..., idx]).min (),= plt.get_cmap("rainbow" ),= 0.5 ,* TG[..., idx].numpy(),= 1 ,= 1 ,= plt.get_cmap("rainbow" ),= 0.0 ,"Δα (radians)" )"Δγ (radians)" )= fig.add_subplot(2 , 3 , 3 , projection= "3d" )* PG[..., idx].numpy(),= "z" ,= (multiplier * PG[..., idx]).min (),= plt.get_cmap("rainbow" ),= 0.5 ,* PG[..., idx].numpy(),= 1 ,= 1 ,= plt.get_cmap("rainbow" ),= 0.0 ,"Δβ (radians)" )"Δγ (radians)" )# Angles = torch.meshgrid(xs, ys, indexing= "ij" )= torch.meshgrid(xs, zs, indexing= "ij" )= torch.meshgrid(ys, zs, indexing= "ij" )= fig.add_subplot(2 , 3 , 4 , projection= "3d" )* XY[..., idx],= "z" ,= (multiplier * XY[..., idx]).min (),= plt.get_cmap("rainbow" ),= 0.5 ,* XY[..., idx].numpy(),= 1 ,= 1 ,= plt.get_cmap("rainbow" ),= 0.0 ,"ΔX (mm)" )"ΔY (mm)" )= fig.add_subplot(2 , 3 , 5 , projection= "3d" )* XZ[..., idx].numpy(),= "z" ,= (multiplier * XZ[..., idx]).min (),= plt.get_cmap("rainbow" ),= 0.5 ,* XZ[..., idx].numpy(),= 1 ,= 1 ,= plt.get_cmap("rainbow" ),= 0.0 ,"ΔX (mm)" )"ΔZ (mm)" )= fig.add_subplot(2 , 3 , 6 , projection= "3d" )* YZ[..., idx].numpy(),= "z" ,= (multiplier * YZ[..., idx]).min (),= plt.get_cmap("rainbow" ),= 0.5 ,* YZ[..., idx].numpy(),= 1 ,= 1 ,= plt.get_cmap("rainbow" ),= 0.0 ,"ΔY (mm)" )"ΔZ (mm)" )return fig, axs